Quantitative ultrasound muscle atrophy evaluation

We developed deep learning methods for quantitative muscle atrophy evaluation using ultrasound images.
Research background
Muscle atrophy due to weightlessness is one of the major health problems encountered by astronauts during medium- and long-term space missions. Prevention and treatment of muscle atrophy in weightlessness is an important guarantee for the successful completion of manned space missions, especially for deep space exploration missions, which demands accurate muscle atrophy evaluation models and related technologies. Among the existing imaging modalities, ultrasonography has the advantages of being portable, non-radioactive, and non-invasive, making it more suitable for evaluating muscle atrophy in special environments such as aerospace. In this project, we have developed radiomics analysis and deep learning methods for quantitative muscle atrophy evaluation using ultrasound images.
The studies were conducted in collaboration with Professor Jianzhong Guo of Shaanxi Normal University and Dr. Xiaoping Chen of the National Key Laboratory of Human Factors Engineering at the China Astronaut Research and Training Center.
- Ultrasound radiomics for longitudinal evaluation of muscle atrophy
This study demonstrates for the first time the capability of radiomics analysis in the evaluation of muscle atrophy using ultrasound images. The stability and informativeness of ultrasound radiomics features were evaluated through a two-stage feature selection procedure. An adaptive longitudinal feature selection and grading network was developed for muscle atrophy evaluation. The results demonstrated the necessity and feasibility of constructing a graded model by fusing multi-stage models in the longitudinal evaluation of muscle atrophy.
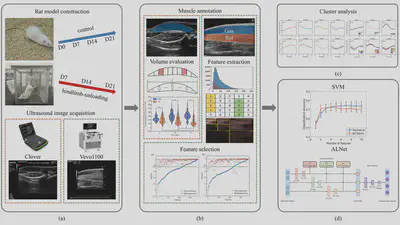
Analysis workflow. (a) Development of hindlimb unloading (HU) rat model and acquisition of ultrasound images. (b) Construction of muscle volume calculation model, extraction and selection of radiomics features. (c) Clustering analysis of ultrasound image radiomics features. (d) Construction of muscle atrophy grading models based on SVM and neural networks.
Read our paper for more details:
Yue Zhang, Getao Du, Yonghua Zhan, Kaitai Guo, Yang Zheng, Liang Tang, Jianzhong Guo, and Jimin Liang*, Muscle Atrophy Evaluation via Radiomics Analysis using Ultrasound Images: A Cohort Data Study, IEEE Transactions on Biomedical Engineering, 69(10):3163-3174, 2022.
- Contrastive disentanglement for quantitative ultrasound muscle atrophy evaluation
In our previous work, we have developed a muscle atrophy grading model using the timestamps as the evaluation criterion, with relatively positive results. However, muscle atrophy is a complex physiological remodeling process characterized by molecular, metabolic, structural, and functional changes. Such complex physiological changes lead to nonlinear time-varying characteristics of the muscle ultrasound image features. Consequently, the use of hind-limb unloading time as a benchmark for the degree of muscle atrophy is inaccurate, which complicates the development of models for quantitative assessment of muscle atrophy and should only be used as a last resort.
In this study, we proposed a new ultrasound imaging-based method for quantifying muscle atrophy that utilizes weakly supervised information between multiple data partitions with controlled variance components, overcoming the limitations of using the weightlessness time as a criterion. We developed a group-supervised contrastive disentanglement network (GCDNeT) to disentangle the individual variances, muscle growth and atrophy components of ultrasound images, and quantify the degree of atrophy using the disentangled atrophy component. The feasibility of GCDNet is validated by the separability, independence, and representativeness of the disentangled components. To simplify the application of GCDNet, a muscle atrophy scoring network requiring no reference images is developed by distilling the GCDNet’s knowledge of muscle atrophy quantization. The results showed that the trend of muscle atrophy scores computed by the atrophy components is similar to that of atrophy at the molecular level. The strength of the proposed methodology allows us, for the first time to our knowledge, to study the muscle growth attribute during hind-limb unloading and the spatial distribution of muscle atrophy.
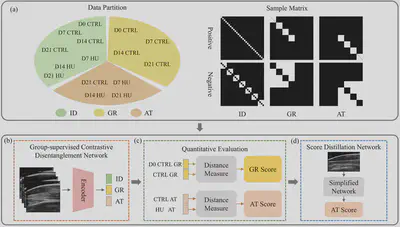
Analysis workflow. (a) Construction of three data partitions with known individual (ID), muscle growth (GR), and muscle atrophy (AT) variance components. Positive and negative samples in each data partition are controlled by the sampling matrix. (b) Group-supervised contrastive disentanglement network (GCDNet) for disentangled representation learning of the IR, GR, AT, and residual (RE) components. (c) Quantitative evaluation of muscle growth and muscle atrophy. (d) Distillation network for simplified application of muscle atrophy evaluation.
Read our paper for more details:
Yue Zhang, Yonghua Zhan, Kaitai Guo, Yang Zheng, Liang Tang, Jianzhong Guo, Jimin Liang*, Contrastive Disentanglement for Quantitative Ultrasound Muscle Atrophy Evaluation, IEEE Transactions on Biomedical Engineering, 71(8):2352-2366, 2024.
- Self supervised temporal ultrasound reconstruction for muscle atrophy evaluation
In this work, we proposed a self-supervised temporal ultrasound reconstruction method based on masked autoencoder to explore the dynamic process of muscle atrophy. A score-position embedding was designed to realize the quantitative evaluation of muscle atrophy. Ultrasound images of the hind limb muscle of six macaque monkeys were acquired consecutively during 38 days of head-down bed rest experiments. Given an ultrasound image sequence, an asymmetric encoder-decoder structure was used to reconstruct the randomly masked images for the purpose of modelling the dynamic muscle atrophy process. We have demonstrated the feasibility of using the position indicator as muscle atrophy score, which can be used to predict the degree of muscle atrophy. This study achieves the quantitative evaluation of muscle atrophy in the absence of accurate evaluation criteria for muscle atrophy.
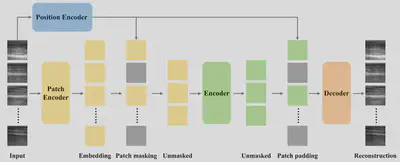
Masked autoencoder on temporal ultrasound images. The input patches are embedded by the patch encoder. Position embeddings from position encoder are added to the embedded patches. The encoder operates on the set of visible patch embeddings. The decoder then processes the full set of encoded patch embeddings and mask tokens to reconstruct the input.
Read our paper for more details:
Yue Zhang, Getao Du, Yonghua Zhan, Kaitai Guo, Yang Zheng, Jianzhong Guo, Xiaoping Chen, and Jimin Liang, Self supervised temporal ultrasound reconstruction for muscle atrophy evaluation, 6th Chinese Conference on Pattern Recognition and Computer Vision, PRCV2023, Xiamen, China, October 13–15, 2023.